VirusTaxo: Taxonomic classification of viruses from the genome sequence using k‑mer enrichment
Abstract
Classification of viruses into their taxonomic ranks (e.g., order, family, genus) provides a framework to organize an abundant population of viruses. Next-generation metagenomic sequencing technologies lead to a rapid increase in generating sequencing data of viruses which require bioinformatics tools to analyze the taxonomy. Many metagenomic taxonomy classifiers have been developed to study microbiomes, but it is particularly challenging to assign the taxonomy of diverse virus sequences and there is a growing need for dedicated methods to be developed that are optimized to classify virus sequences into their taxa. VirusTaxo, developed using diverse (e.g., 402 DNA and 280 RNA) genera of viruses, has an average accuracy of 93% at genus level prediction in DNA and RNA viruses. VirusTaxo outperformed existing taxonomic classifiers by assigning taxonomy to a larger fraction of metagenomic contigs compared to other methods. Benchmarking of VirusTaxo on a collection of SARS-CoV-2 sequencing libraries and metavirome datasets suggests that VirusTaxo can characterize virus taxonomy from highly diverse contigs and provide a reliable decision on the taxonomy of viruses.
Keywords: Virus Taxonomy, Hierarchical Classification, k-mer, Genome
Results:
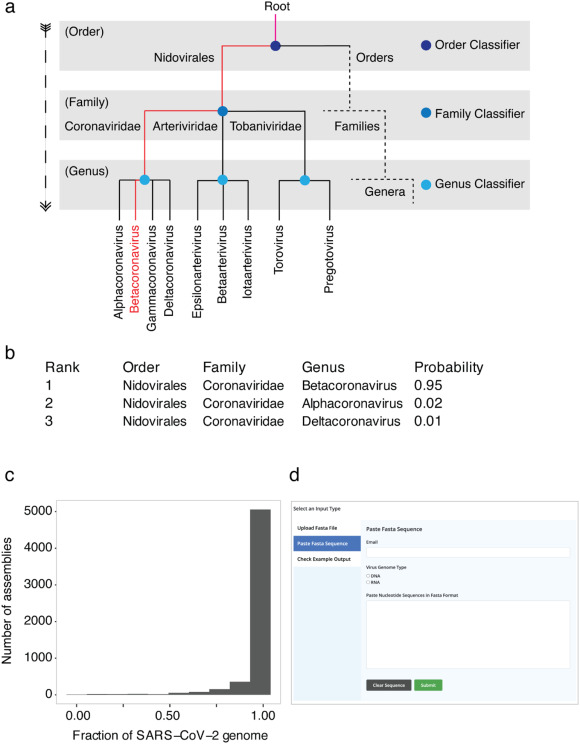
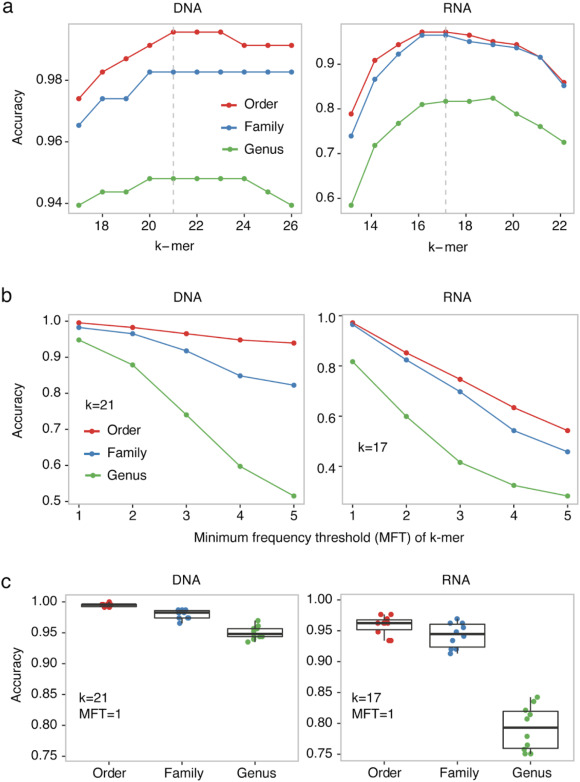
Classification of taxonomic ranks of viruses using VirusTaxo
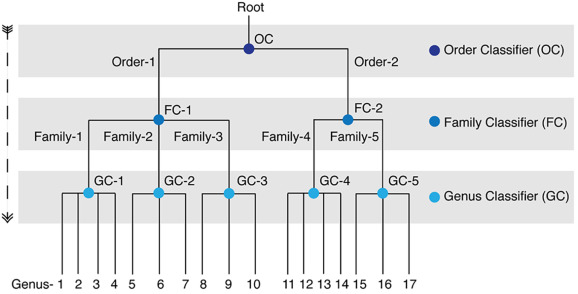
Accuracy of VirusTaxo for order, family, and genus level classification in the pilot dataset.
Benchmarking of VirusTaxo for SARS-CoV-2 genomes